Extract-Transform-Load
Performing ETL activities consume a significant part of the average data science workflow. With Akumen, we aim to streamline it somewhat by using PETL - a Python ETL library and makes transforms relatively simple. This model is simple, because there isn’t much ETL required for this dataset - it’s all basically in the format that we need. Hence, we just perform some minor transformations to show the workflow.
To create an ETL model, you can do the following:
- Go to the App Manager, and select
Create Application -> Python Model, namedETL - Breast Cancer. - To use PETL, we need to include it in the build environment. Create a new file called
Dockerfile(which will be created from a template) and add thepetlinstall line as below:
FROM quay.io/optika.solutions/akumen-python:3.7.2
RUN pip3 install petl- Clear out
main.pyand add the following, step-by-step:
import petl
from akumen_api import get_resultsThe first line imports petl, the Python ETL library. The second line imports a helper from Akumen’s API functions, get_results, which retrieves data from another model’s execution. We’ll use this to get the data from our data connector in the previous section.
def akumen(**kwargs):
"""
Parameters:
- Input: data [scenario]
- Input: view [string]
- Input: scope [string]
- Output: results [tabular]
"""
print('Running Akumen model...')This section is our standard Akumen wrapper function and parameter definitions. In this model, we take three parameters:
data- a scenario input that points to our Connector model. We use this withget_resultsto retrieve the connector’s result data (which is our breast cancer data).view- the name of the view that we want to retrieve results from. For our connector, this isdata_vw.scope- the scope level that we want to retrieve data for. We’re only retrieving data from a single scenario, soscenarioscope is fine.
# grab the whole resultset
input = petl.fromdataframe(
get_results(kwargs.get('data'), kwargs.get('view'), kwargs.get('scope'))
)Using get_results, we retrieve the dataset from the Connector model and pass it into petl. get_results returns a dataframe, which is easily imported into petl for modification.
# reflow diagnosis into a 0/1 value, since autosklearn doesn't operate on labels
input = petl.convert(input, 'diagnosis', lambda v: 0 if v == 'B' else 1)As a simple modification, we reform the diagnosis column into a binary integer format. petl provides a large number of transformation functions that can be used, but other libraries or code can be used here too - fuzzy matching, integration, or others.
# strip off the akumen columns so we don't duplicate them
for col in ['studyname', 'scenarioname', 'id']:
input = petl.cutout(input, col)We also want to cut off some Akumen-standard columns, so they’re not duplicated by this model.
# or we can return the df directly to the `return` of the `akumen()` function:
return {
'results': petl.todataframe(input)
}And we return the resulting dataframe for output.
The entire resulting file is below:
import petl
from akumen_api import get_results
def akumen(**kwargs):
"""
Parameters:
- Input: data [scenario]
- Input: view [string]
- Input: scope [string]
- Output: results [tabular]
"""
print('Running Akumen model...')
# grab the whole resultset
input = petl.fromdataframe(
get_results(kwargs.get('data'), kwargs.get('view'), kwargs.get('scope'))
)
# reflow diagnosis into a 0/1 value, since autosklearn doesn't operate on labels
input = petl.convert(input, 'diagnosis', lambda v: 0 if v == 'B' else 1)
# strip off the akumen columns so we don't duplicate them
for col in ['studyname', 'scenarioname', 'id']:
input = petl.cutout(input, col)
# or we can return the df directly to the `return` of the `akumen()` function:
return {
'results': petl.todataframe(input)
}For inputs, you should use:
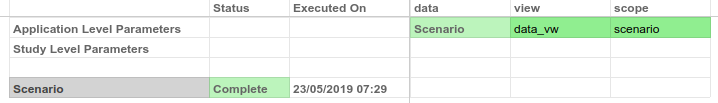
data: the connector model we previously built.view:data_vwscope:scenario
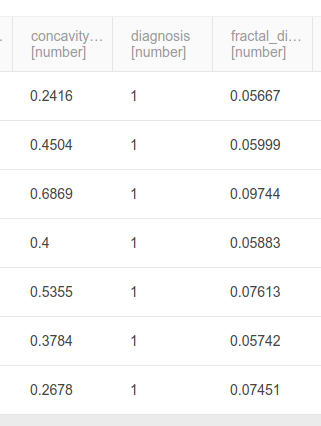
Execute the model and go to the data tab, and you should see that the diagnosis column is now 0/1 instead of B/M.